About
Mutation-induced protein thermodynamic stability changes (DDG) are crucial for understand protein biophysics, genomic variant interpretation, and mutation-related diseases. We introduce SCpre-seq, a residue level 3D structure-based prediction tool to assess the effects of single point mutation on protein thermodynamic stability and applying to dingle-domain monomeric proteins. Given protein sequence with single mutations as the input, the proposed model integrated both sequence-level features of mutant residues and mutation-based structure features. Our stability predictor outperformed previously published methods on various benchmarks. SCpre-seq will be a dedicated resource for assessing both somatic and germline substitution mutations in biological and medical research on genomics and proteomics.
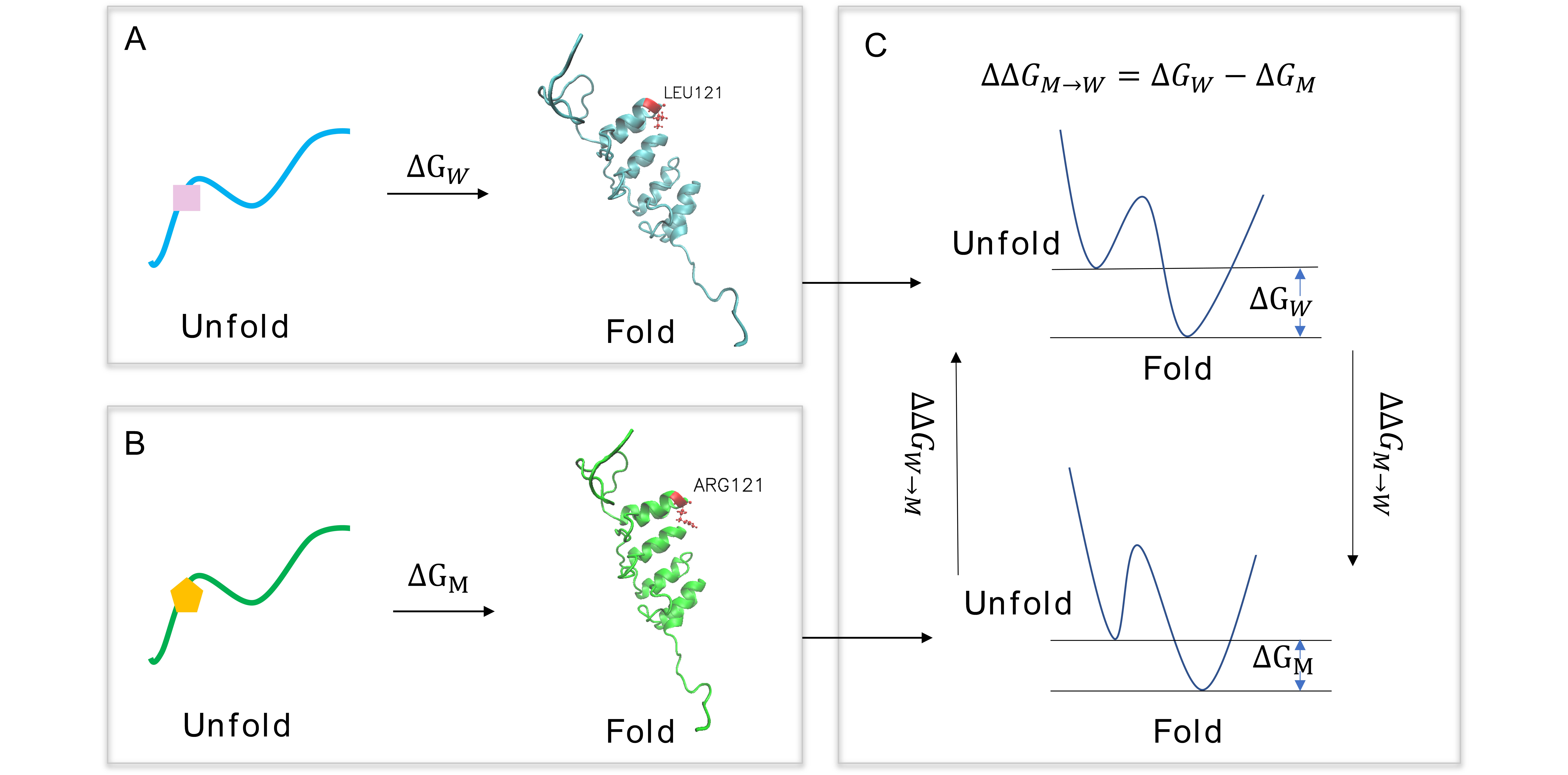
What is DDG?
Protein thermodynamic stability changes of single point mutation are changes of the Gibbes free energy for the biophysical process of protein folding between two states before and after single point mutation on the protein[1]. A quantified change of Gibbs free energy of a protein between the folding and unfolding status is usually represented as DG. When a point mutation is present and a residue is substituted in a protein, the original protein would be a “reference state”, likely called “wild-type protein”. The protein mutated is called “mutation protein”.