Sequence-based stability changes prediction
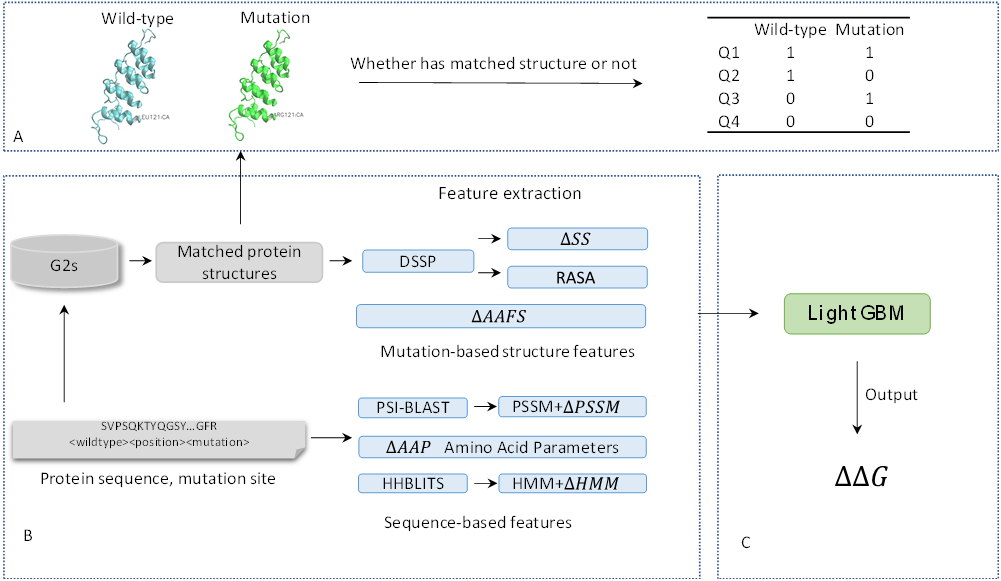
SCpre-seq, a residue level 3D structure-based prediction tool to assess single point mutation effects on protein thermodynamic stability and applying to dingle-domain monomeric proteins. Given protein sequence with single mutations as the input, the proposed model integrated both sequence level features of mutant residues and residue level mutation-based 3D structure features.
SCpre-seq’s applications
SCpre-seq can be applied to predicting stability changes on single monomeric proteins which tertiary structures are unavailable.
SCpre-seq can be used to assess both somatic and germline substitution mutations such as p53 in biological and medical research on genomics and proteomics.
Mutation-based background knowledge of variants obtained from G2S which surveys the whole mutations in PDB can be used to makes up for the shortage of structural information for sequence-based methods.
A reliable mutation-based residue-level 3D structure information has applied to improve the model’s performance.
Reference
ACKNOWLEDGEMENTS
Support
Feel free to submit an issue or send us an email. Your help to improve SCpre-seq is highly appreciated.