Mutation structure preparation
[G2S](https://g2s.genomenexus.org) provides a real-time web API that provides an automated mapping of genomic variants on 3D protein structures. Giving protein sequence and the position of a variant as the query, G2S searches to get a 3D structure profile that is a list of protein structures with active residues whose positions are matched with the position of input variant after aligning with queried sequence. We divided this list of protein structures of variants into two classes based on the type of the wild-type and mutant amino acids from the input data (Supplemental Figure S1). Due to the availabilities of the PDB structures, four categories of PDB structures are adopted. Q1: Both tertiary structures in the 3D structure profile whose active residues are matched to wild-type residue and tertiary structures in the 3D structure profile whose active residues are matched to mutant residue are available. Q2: Tertiary structures in the 3D structure profile whose active residues are wild-type residue are available while structures which matched mutant residue are unavailable. Q3: Tertiary structures in the 3D structure profile whose active residues are wild-type residue are unavailable while tertiary structures which matched mutant residue are available. Q4: Neither tertiary structure in 3D structure profile which matched wild-type residue nor tertiary structures which matched mutant residue are available.
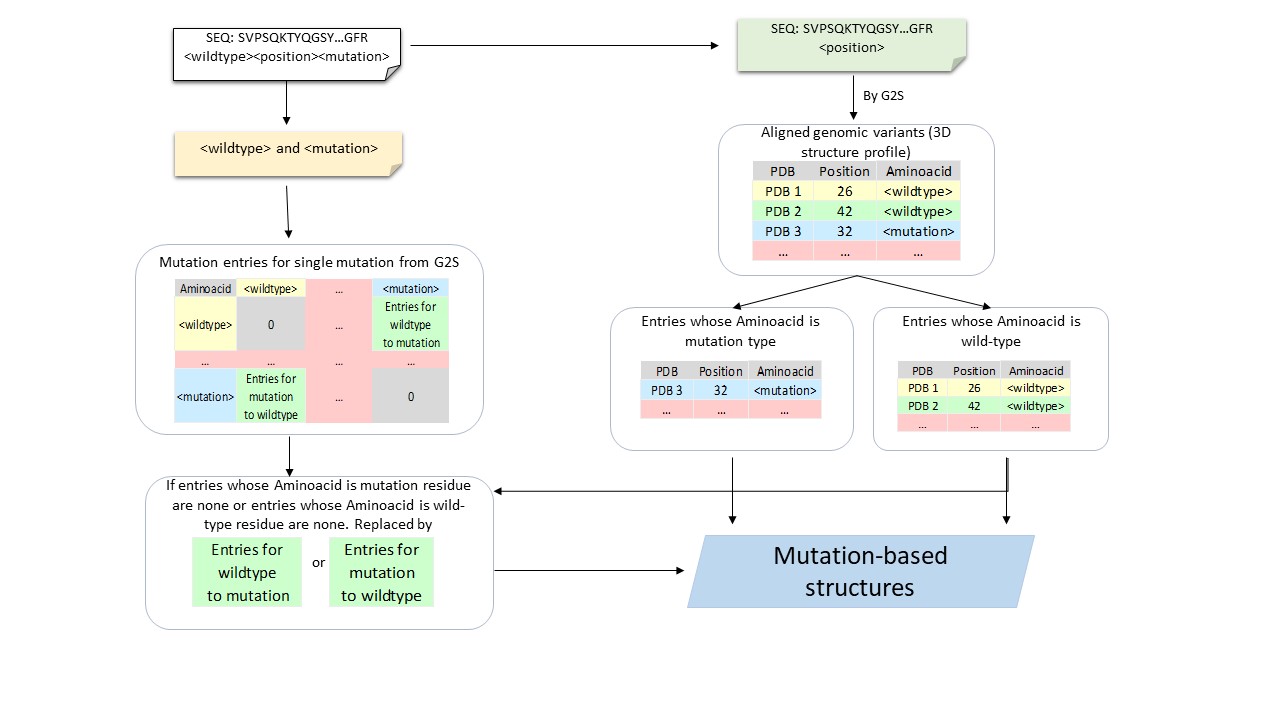
The samples count for 4 situations when mutation-based structures are available or not on datasets S1676, S543 and S236.
Dataset |
Q1 |
Q2 |
Q3 |
Q4 |
|---|---|---|---|---|
S543 |
172 |
366 |
1 |
4 |
S236 |
46 |
159 |
0 |
31 |
S1676 |
706 |
950 |
0 |
20 |